RuleBender: integrated modeling, simulation and visualization for rule-based intracellular biochemistry
RuleBender: integrated modeling, simulation and visualization for rule-based intracellular biochemistry
Smith, A.M., Xu, W., Sun, Y., Faeder, J.R., Marai, G.E.
- Location: Providence, RI
- Link: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3355338/pdf/1471-2105-13-S8-S3.pdf
- PDF: rulebender2011.pdf
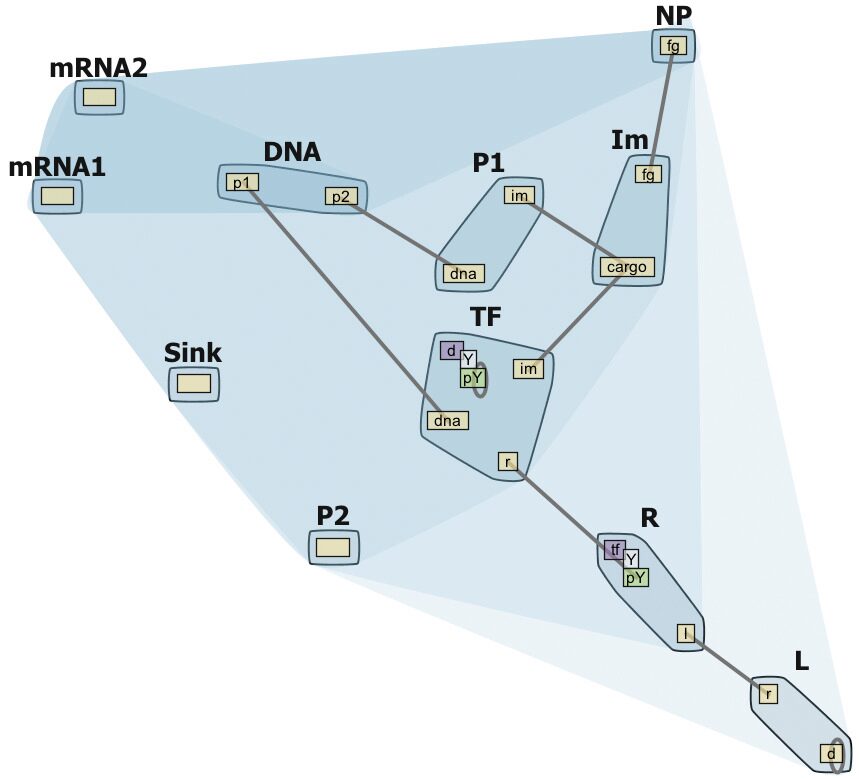
- Caption: Contact Map with molecule compartment hierarchy (extracellular, cytoplasmic, nucleus etc).
Background: Rule-based modeling (RBM) is a powerful and increasingly popular approach to modeling cell signaling networks. However, novel visual tools are needed in order to make RBM accessible to a broad range of users, to make specification of models less error prone, and to improve workflows.
Results: We introduce RuleBender, a novel visualization system for the integrated visualization, modeling and simulation of rule-based intracellular biochemistry. We present the user requirements, visual paradigms, algorithms and design decisions behind RuleBender, with emphasis on visual global/local model exploration and integrated execution of simulations. The support of RBM creation, debugging, and interactive visualization expedites the RBM learning process and reduces model construction time; while built-in model simulation and results with multiple linked views streamline the execution and analysis of newly created models and generated networks.
Conclusion: RuleBender has been adopted as both an educational and a research tool and is available as a free open source tool at http://www.rulebender.org. A development cycle that includes close interaction with expert users allows RuleBender to better serve the needs of the systems biology community